Pathogens in Focus
The two model organisms studied in Cynthia Sharma’s lab are the widespread human pathogens Helicobacter pylori, carried by around half of the world's population and a cause of gastritis and stomach cancer, and the related bacterium Campylobacter jejuni, which is currently the most common cause of bacterial food poisoning.
In June 2010, she moved from the Max Planck Institute for Infection Biology in Berlin to the Research Center for Infectious Diseases (ZINF) at the University of Würzburg in order to establish her own laboratory as the head of an independent ZINF young investigator research group. Today, Sharma heads the Chair of Molecular Infection Biology II at the University of Würzburg, which was newly established in 2017, and has been the ZINF's spokesperson since 2018. In addition, she has been the mother of a daughter since 2016.
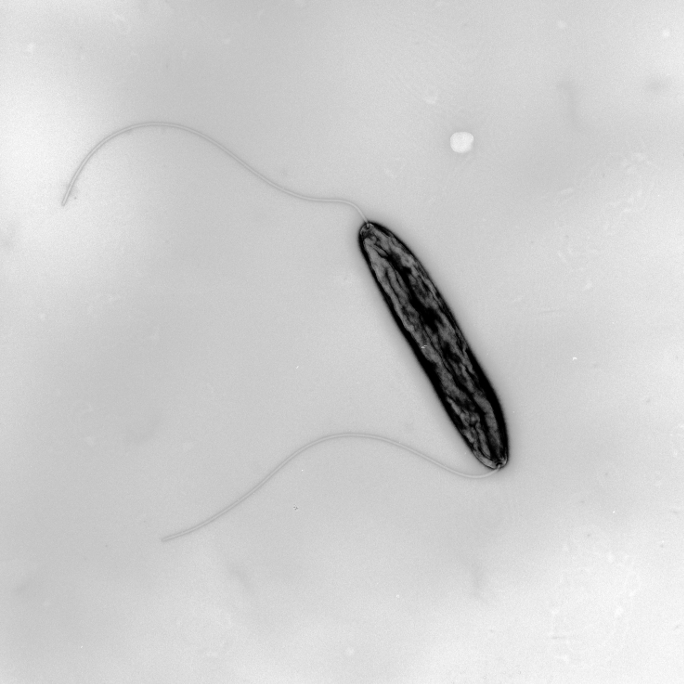
Sharma is primarily interested in the molecular biological processes that regulate genes involved in virulence in bacterial pathogens, with a focus on using global genomics and transcriptomics techniques combined with molecular genetics and biochemistry. In 2009, together with her doctoral supervisor Jörg Vogel and scientists from Leipzig and Bordeaux, she established a new high-throughput RNA-sequencing approach, which can map gene start sites genome-wide (Sharma et al., Nature, 2010; PMID: 20164839). This approach also revealed the presence of an unexpected wealth of small regulatory RNA molecules (sRNAs) in H. pylori. Such sRNAs are an emerging class of central regulators in the bacterial stress response or virulence control.
Research on Mechanisms of Bacterial Gene Regulation, Stress Response, and Pathogenicity
From 1998 to 2004, Sharma studied biology with a focus on biophysics and bioinformatics at the University of Düsseldorf. She pursued her doctoral studies at the Max Planck Institute for Infection Biology (MPIIB) in Berlin (2004 to 2009) and also visited the University of Bordeaux for a short research stay during her doctoral studies. After briefly working as a postdoc at the MPIIB and the National Institutes of Health (USA), she moved to the University of Würzburg in 2010. In 2016, she was appointed interim chair of the Chair of Molecular Infection Biology II, which she has been leading as full professor since 2017.
How do bacterial pathogens adapt to changing environments in order to survive and cause disease, and how can we use the knowledge of these mechanisms to develop new antimicrobial strategies?Prof. Cynthia Sharma, Spokesperson of the Research Center for Infectious Diseases, University of Würzburg
Sharma has received several awards for her research—including the Heinz Maier-Leibnitz Prize of the German Research Foundation (DFG) in 2015. Moreover, she was a member of the Young Academy of the Bavarian Academy of Sciences from 2012 to 2017.
Today, Cynthia Sharma has further expanded her scientific research focus on bacterial gene regulation and virulence control. Exploring so-called small proteins or the bacterial CRISPR-Cas system and the development of new three-dimensional infection models based on tissue engineering are only a few examples of the Sharma Group's focal areas of research.
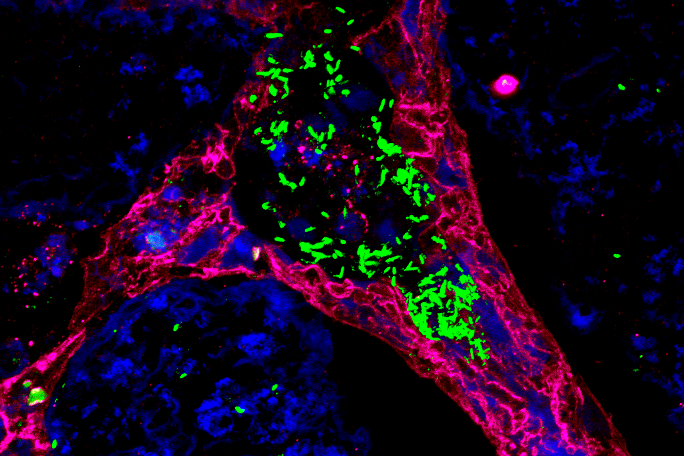
Within the framework of the recently established Bavarian research network bayresq.net, which aims to develop new strategies against multi-drug-resistant pathogens by means of digital networking, Sharma closely collaborates with her project partners in their "StressRegNet" consortium: junior research group leader Ana Rita Brochado from the Biocenter/ZINF in Würzburg and Professor Christian Müller from the LMU Munich. Their collaborative, interdisciplinary project aims to expose the bacterial pathogens Salmonella and Campylobacter to more than 3,000 different host-specific signal molecules and measure how different genes related to antibiotic resistance and stress responses respond using automated high-throughput technologies. By application of machine learning methods to the generated data, regulatory networks will be inferred to provide insight into how each pathogen responds to specific signals and adapt to changing environments in the host or to antibiotics. Their overarching research question is: "How do bacterial pathogens adapt to changing environments in order to survive and cause disease, and how can we use the knowledge of these mechanisms to develop new antimicrobial strategies?", Sharma says.
New Technologies Provide Novel Insights
Research on RNA molecules is one of the current main areas of research in Würzburg at the Research Center for Infectious Diseases (ZINF), at the Institute for Molecular Infection Biology (IMIB), as well as at the Helmholtz Institute for RNA-based Infection Research (HIRI). Global transcriptome analyses provide information about the expressed RNA molecules inside cells and help to reveal non-coding RNA molecules that play key roles during infection. "The knowledge about functions of RNA molecules in the infection process and the regulatory pathways they control can contribute to developing new antimicrobial strategies," Sharma says.
In the future, researchers will increasingly focus on the interaction of different regulatory networks in both the pathogen and the host as well as on the individual host cell level instead of across the whole cell population: "Single cell sequencing enables us to obtain a detailed understanding of how heterogeneity of pathogen and host cells underlies different processes during infections," Sharma says. What is the host target cell for a given pathogen? Which niches does a pathogen occupy in the body? How does the immune system react to this attack? All of these questions can be approached using single-cell analysis.
Career at the Research Center for Infectious Diseases (ZINF)
The Research Center for Infectious Diseases (ZINF) at the University of Würzburg is dedicated to the study of infectious diseases, and in particular of the molecular principles of host-pathogen interactions. It provides junior research group leaders, postdocs, and PhD students a highly interdisciplinary and international environment in the field of infectious disease research.
Interdisciplinary and Inter-Faculty Research
The ZINF is a central, faculty-spanning scientific institution of the University of Würzburg, where experts in microbiology, parasitology, virology, immunology, chemistry, pharmacy, and clinicians join forces to address different questions in infection biology.
An Ideal Stepping Stone in the Development of Early-Career Researchers
Promoting young research talents is one of the ZINF's central aspects. Heading a junior research group at ZINF offers young scientists the opportunity to establish themselves in the research world with their own topic, usually for a period of five years. Since the foundation of ZINF in 1993, about 30 young scientists from Germany and abroad have been recruited to Würzburg to set up their own independent laboratory after their postdoctoral research.
Research Topics of Current Junior Research Groups
- Organoids as new infection models (Research Group Bartfeld)
- The role of microbiota in fungal infections (Research Group Perez)
- Regulatory RNA molecules in anaerobic pathogens and in the microbiome (Research Group Faber)
- High-throughput technologies for investigating effects of antibiotic combinations (Research Group Brochado)
Former ZINF junior research group leaders have been appointed to national or international associate or full professorships or research group leader positions and are now continuing their work at universities in, e.g., Munich, Heidelberg, Konstanz, Kiel, or Madrid.
The Future is Digital
New technologies have great potential to provide new insights, but they also generate enormous and complex datasets. Their evaluation and interpretation will be more and more important for infection biologists like Sharma. New analysis tools are required for this. "New methods for data analysis and visualization are required to understand what is happening in millions of cells and to decipher the interaction of different regulatory networks, not only within cells but also between host and pathogen," Sharma explains.
She is therefore convinced that deep learning, artificial intelligence, and machine learning approaches will shape the future analyses of big datasets in infection research. For example, researchers have already developed algorithms that can use global air passenger data to model the distribution and origin of a pathogen across continental borders, a tool that could be especially important in times of the coronavirus pandemic. The same computational strategies could also be used, for example, to monitor and model signaling cascades in a cell triggered by a pathogen.
More about the University of Würzburg



